Understanding the Techniques and Differentiating qPCR from RT-PCR
Feb 13, 2024
Polymerase Chain Reaction (PCR), also known as PCR amplification, is a technique used to amplify specific DNA fragments, allowing for exponential growth in the number of DNA segments. Before the advent of PCR technology, researchers had to handle nucleic acid samples with extreme care because each DNA sample was precious. With the emergence of PCR technology, this limitation was overcome. The principle of PCR is similar to DNA replication in vivo, and it can be seen as a special DNA replication outside the organism. With PCR technology, even a tiny amount of DNA can be amplified as long as it is separated from the sample.
The PCR process can be roughly divided into 3 steps: Denaturation—Annealing—Extension. Denaturation (90°C-96°C): Double-stranded DNA templates break hydrogen bonds under heat to form single-stranded DNA. Annealing (60°C-65°C): The system temperature decreases, and primers bind to the DNA template, forming local double strands. Extension (70°C-75°C): With the action of Taq polymerase (optimal activity at around 72°C), using dNTP as raw materials, extension starts from the 3′ end of the primer in the 5′→3′ direction, synthesizing a DNA chain complementary to the template.
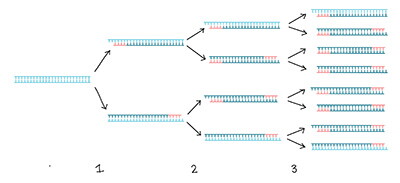
After amplification, we need to detect the amplified DNA. Conventional PCR techniques usually rely on gel electrophoresis. Gel electrophoresis separates DNA based on differences in molecular weight, enabling identification and purification of DNA. Gel electrophoresis can determine whether the size of the amplified DNA fragment band matches the desired band size, identify whether there are impurities, assess the specificity of amplification, and evaluate primer design quality.
Regarding qPCR and Real-time PCR: Many people mistakenly think that RT-PCR is an abbreviation for Real-time PCR, thus confusing the two. However, they are different. We'll talk about RT-PCR later.
Real-time PCR, also known as quantitative PCR (qPCR), is what many are familiar with, and they are indeed the same thing. In the detection methods mentioned after PCR, gel electrophoresis is usually employed to analyze the products. Real-time PCR, however, utilizes fluorescent chemicals to detect DNA amplification during the PCR process, eliminating the need for gel electrophoresis and greatly simplifying the experimental steps.
The experimental principle of qPCR: In simple terms, qPCR introduces fluorescent groups throughout the amplification process. As the number of DNA copies increases exponentially, the fluorescence signal in the reaction system also enhances. By detecting the strength of the fluorescence signal, quantitative analysis of DNA amplification can be performed.
Currently, depending on different fluorescent substances, qPCR can be roughly divided into two types. One uses fluorescent dyes, mainly SYBR Green I dye, which binds to double-stranded DNA and emits fluorescence when bound, almost undetectable without binding, with fluorescence signal enhancing with the increase of copy number. The other type uses fluorescent probes, also known as TaqMan probe method. Its principle is to design a fluorescent probe that specifically binds to the target gene. The probe contains two groups: a fluorescent group and a quencher group. Normally, the quencher group prevents the fluorescent group from emitting fluorescence. During amplification, when the probe binds to the template DNA, and when it is amplified to the probe position, the two groups are separated, allowing the fluorescent group to emit fluorescence. Similarly, with the increase of copy number, the fluorescence signal enhances.
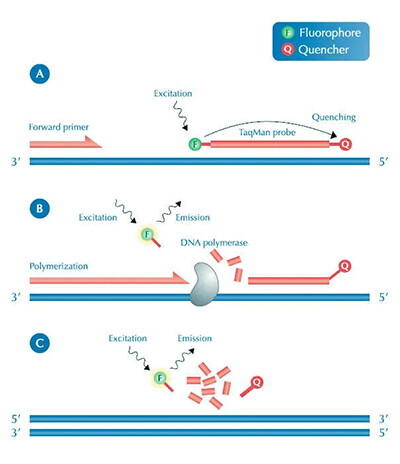
With this quantitative relationship, PCR amplification curves can be plotted. The curve shows the fluorescence signal changing with the number of cycles, allowing analysis of PCR conditions. The ideal curve should exhibit an "S" shape growth, but the analysis of the curve is relatively complex, and I won't go into much detail here. At least we know that with this technology, we don't have to laboriously run gel electrophoresis after each PCR, and we can analyze our PCR results more accurately. Therefore, this technology is increasingly widely used. It's worth mentioning that in the current pandemic situation, when determining whether a tested individual is negative or positive after nucleic acid sampling, qPCR technology is used. If a fluorescence signal can be detected after a certain number of cycles of amplification, we can consider the tested individual as positive; otherwise, negative. Regarding RT-PCR: Compared to the former, RT-PCR is much simpler. Its full name is Reverse Transcription PCR, or Reverse Transcriptase PCR. First, we need to understand what reverse transcription is. We generally refer to the process of genetic information flowing from DNA to RNA as transcription. Conversely, the process of synthesizing DNA using RNA as a template is called reverse transcription. The general process of this technology is to first synthesize the corresponding cDNA using RNA as a template under the action of reverse transcriptase, and then perform regular PCR using cDNA as a template. In essence, it adds a reverse transcription process before the PCR process. After this, we can use conventional PCR methods or qPCR for amplification. For example, viruses usually use RNA as genetic material. When conducting pathogen detection, we can only extract their RNA, so reverse transcription is needed. Other PCR techniques: In addition to the PCR techniques introduced above, there are now other techniques such as digital PCR, also known as nucleic acid molecule absolute quantification technology, which quantifies DNA more absolutely than fluorescent quantification; and nested PCR, etc. However, their essence is inseparable from the three steps of PCR mentioned earlier.
Pulse provides corresponding consumables for the PCR process:
PCR tubes/eight-tube strips/plates: Made of polypropylene (PP) material to ensure that the material does not deform or damage during denaturation at high temperatures; thin-walled design for rapid heat transfer during temperature increase, improving PCR efficiency; excellent sealing performance, even with minimal PCR systems, there is no evaporation due to high temperatures, reducing the loss of reaction volume; enzyme-free and heat-resistant, ensuring that your DNA samples do not degrade due to contamination; available in white and transparent colors, white reduces fluorescence refraction in qPCR experiments, obtaining the most sensitive and accurate fluorescence signal; plate-type consumables provide versions with different skirt types to facilitate matching in different PCR instruments or automated applications.
Previous: How to Select Laboratory Pipette Tips
Next: The Development of Micropipettes



