How to Accurately Obtain Experimental Results in PCR
Mar 14, 2024
Quantitative Polymerase Chain Reaction (qPCR) analysis is widely utilized in scientific laboratories due to its high sensitivity, allowing for precise detection and quantification of nucleic acids. However, several factors, including pipetting techniques, can influence the results of qPCR analysis. PCR Master Mix, typically used during qPCR preparation, can be challenging to pipette accurately. In this study, we investigated the use of electronic pipettes for transferring Master Mix in qPCR.
Our research demonstrates that using electronic pipettes with low-retention filter tips or standard filter tips for Master Mix pipetting yields accurate and precise results, ensuring reproducibility of pipetting speed and outcomes. Therefore, we conclude that using electronic pipettes for transferring Master Mix ensures reproducible and reliable results in PCR-based analyses.
Introduction
PCR-based applications have become pivotal in biopharmaceutical processes, clinical diagnostics, and academic research. However, variability in experimental results during quantitative Polymerase Chain Reaction (qPCR) execution can pose challenges, with pipetting errors being a significant source of variation. Pipetting Master Mix reagents during qPCR preparation presents challenges. Master Mix typically consists of polymerase, dNTPs, MgCl2, and buffer solution containing components such as Tween and glycerol.
The aim of this application note was to assess the reliability of using electronic pipettes for transferring Master Mix in PCR analysis applications. During quantitative PCR preparation, electronic pipettes paired with low-adhesion filter tips were used to transfer Master Mix, and the experimental results (quantitative cycles, Cq) were evaluated based on accuracy and precision (reproducibility).
Methods
Electronic pipettes and pipette tips selection involved the use of electronic pipettes, low-adhesion filter tips, and standard filter tips. Multiple dispense mode was employed. During Master Mix transfer, the electronic pipette speed parameter was set to 1. For qPCR preparation, electronic pipettes, filter tips, and low-adhesion filter tips were used. EP tubes were utilized for DNA sample preparation and Master Mix preparation. PCR Master Mix reservoir solution used for all tests comprised MaximaSYBR Green qPCR Master Mix (without ROX), Escherichia coli uidA gene primers, and nuclease-free water. Eight 15μL Master Mix replicate samples were transferred to PCR plate wells for various test conditions using the pipette. No-template control (NTC) samples contained no Escherichia coli genomic DNA and were supplemented with 5μL of nuclease-free water. Similarly, 5μL aliquots of a series of dilution standards containing 1×106, 1×105, 1×104, 1×103, and 1×102 copies/μL of Escherichia coli gDNA were transferred using the pipette. Each test well contained 5μL of Escherichia coli gDNA with 1×103 copies/μL. All DNA samples were added to PCR tubes using the electronic pipette's multiple dispense function and low-adhesion filter tips in the same manner. qPCR was performed using a 480 qPCR instrument.
The cycling parameters were as follows: initial denaturation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 10 seconds, annealing at 55°C for 10 seconds, and extension at 75°C for 15 seconds, with a final extension at 75°C for 10 seconds. Quantification of SYBR green fluorescence excitation in standards, controls, and samples was conducted. Quantitative cycle (Cq) values and actual copy numbers were determined using software, and results were analyzed using MS Excel.
Data Analysis
The percentage system error (%S) of Cq values reflects the error of the instrument system (pipette and tip system) in handling Master Mix. Random error during pipetting is a measure of result precision, reflecting differences between replicate samples in the experiment. The percentage random error (%R) of Cq values reflects reproducibility of results and may be influenced by the experimenter's pipetting technique.
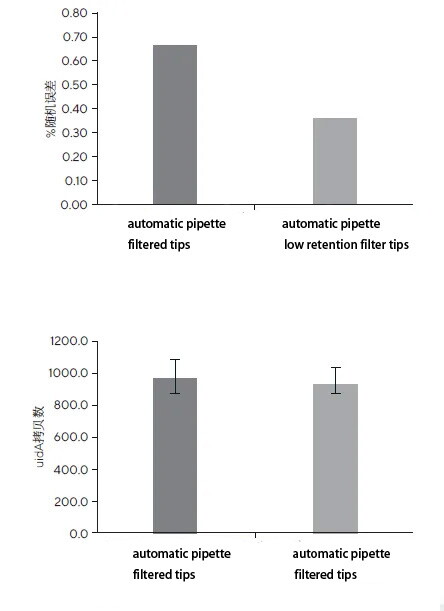
As shown in the figure, when Master Mix was transferred using low-adhesion filter tips with the electronic pipette's multiple dispense mode, the obtained results were as follows: Cq = 24.54 ± 0.09, percentage system error of Cq = 0.12, percentage random error of Cq = 0.37; in comparison, results obtained using standard filter tips were: Cq = 24.52 ± 0.16, percentage system error of Cq = 0.04, percentage random error of Cq = 0.67. In both cases, the results obtained using the electronic pipette demonstrated good reproducibility (percentage random error), with the percentage system error of Cq values maintained at a low level. The electronic pipette's multiple dispense mode ensured sequential distribution of Master Mix to all eight replicate wells after a single aspiration, significantly enhancing pipetting speed while reducing the number of pipette tip changes, thereby promoting environmental sustainability and minimizing differences resulting from tip replacements. Discussion The study demonstrates that using electronic pipettes in qPCR preparation can yield excellent results, with very low percentage error. Compared to standard filter tips, using electronic pipettes in multiple dispense mode with low-adhesion filter tips provides superior result reproducibility (precision) for laboratories still using standard pipette tips for Master Mix transfer.
Our research demonstrates that using electronic pipettes with low-retention filter tips or standard filter tips for Master Mix pipetting yields accurate and precise results, ensuring reproducibility of pipetting speed and outcomes. Therefore, we conclude that using electronic pipettes for transferring Master Mix ensures reproducible and reliable results in PCR-based analyses.
Introduction
PCR-based applications have become pivotal in biopharmaceutical processes, clinical diagnostics, and academic research. However, variability in experimental results during quantitative Polymerase Chain Reaction (qPCR) execution can pose challenges, with pipetting errors being a significant source of variation. Pipetting Master Mix reagents during qPCR preparation presents challenges. Master Mix typically consists of polymerase, dNTPs, MgCl2, and buffer solution containing components such as Tween and glycerol.
The aim of this application note was to assess the reliability of using electronic pipettes for transferring Master Mix in PCR analysis applications. During quantitative PCR preparation, electronic pipettes paired with low-adhesion filter tips were used to transfer Master Mix, and the experimental results (quantitative cycles, Cq) were evaluated based on accuracy and precision (reproducibility).
Methods
Electronic pipettes and pipette tips selection involved the use of electronic pipettes, low-adhesion filter tips, and standard filter tips. Multiple dispense mode was employed. During Master Mix transfer, the electronic pipette speed parameter was set to 1. For qPCR preparation, electronic pipettes, filter tips, and low-adhesion filter tips were used. EP tubes were utilized for DNA sample preparation and Master Mix preparation. PCR Master Mix reservoir solution used for all tests comprised MaximaSYBR Green qPCR Master Mix (without ROX), Escherichia coli uidA gene primers, and nuclease-free water. Eight 15μL Master Mix replicate samples were transferred to PCR plate wells for various test conditions using the pipette. No-template control (NTC) samples contained no Escherichia coli genomic DNA and were supplemented with 5μL of nuclease-free water. Similarly, 5μL aliquots of a series of dilution standards containing 1×106, 1×105, 1×104, 1×103, and 1×102 copies/μL of Escherichia coli gDNA were transferred using the pipette. Each test well contained 5μL of Escherichia coli gDNA with 1×103 copies/μL. All DNA samples were added to PCR tubes using the electronic pipette's multiple dispense function and low-adhesion filter tips in the same manner. qPCR was performed using a 480 qPCR instrument.
The cycling parameters were as follows: initial denaturation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 10 seconds, annealing at 55°C for 10 seconds, and extension at 75°C for 15 seconds, with a final extension at 75°C for 10 seconds. Quantification of SYBR green fluorescence excitation in standards, controls, and samples was conducted. Quantitative cycle (Cq) values and actual copy numbers were determined using software, and results were analyzed using MS Excel.
Data Analysis
The percentage system error (%S) of Cq values reflects the error of the instrument system (pipette and tip system) in handling Master Mix. Random error during pipetting is a measure of result precision, reflecting differences between replicate samples in the experiment. The percentage random error (%R) of Cq values reflects reproducibility of results and may be influenced by the experimenter's pipetting technique.

As shown in the figure, when Master Mix was transferred using low-adhesion filter tips with the electronic pipette's multiple dispense mode, the obtained results were as follows: Cq = 24.54 ± 0.09, percentage system error of Cq = 0.12, percentage random error of Cq = 0.37; in comparison, results obtained using standard filter tips were: Cq = 24.52 ± 0.16, percentage system error of Cq = 0.04, percentage random error of Cq = 0.67. In both cases, the results obtained using the electronic pipette demonstrated good reproducibility (percentage random error), with the percentage system error of Cq values maintained at a low level. The electronic pipette's multiple dispense mode ensured sequential distribution of Master Mix to all eight replicate wells after a single aspiration, significantly enhancing pipetting speed while reducing the number of pipette tip changes, thereby promoting environmental sustainability and minimizing differences resulting from tip replacements. Discussion The study demonstrates that using electronic pipettes in qPCR preparation can yield excellent results, with very low percentage error. Compared to standard filter tips, using electronic pipettes in multiple dispense mode with low-adhesion filter tips provides superior result reproducibility (precision) for laboratories still using standard pipette tips for Master Mix transfer.
Previous: Introduction of Automated Pipetting
Next: Single-Channel Pipettes and Multi-Channel Pipettes



